genomics
What is genomics?
Genomics is the study of an organism's entire genome, including all its genes and how those genes interact with each other and their environment. The genome contains the organism's complete set of DNA and is embedded in nearly every cell of that organism.
Genomics attempts to understand all aspects of the genome's function, organization, evolution and other characteristics. It's a rapidly growing field that's playing an ever-increasing role in healthcare, although it's also finding applications in agriculture, biotechnology, anthropology and other social sciences.
What is DNA?
Genomics is concerned primarily with an organism's DNA, the basic building block of the genome. DNA is a hereditary, chemical compound that carries instructions for the organism's development and function. It is made up of two linked strands that twist around each other to form a double helix.
The DNA strands connect to each other through four nucleotide bases: adenine (A), cytosine (C), guanine (G) and thymine (T). The nucleotides bind the strands together like rungs on a ladder, with each rung made up of two nucleotides. The nucleotides always bind in specific pairs: adenine with thymine and cytosine with guanine.
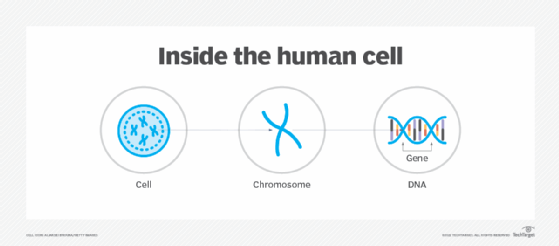
Certain sections of the DNA are segmented into genes, which carry instructions for producing the proteins needed to build and repair tissues and organs. In humans, genes determine individual characteristics like eye color or height, although they can also be responsible for certain diseases and disabilities.
Most of an organism's DNA and genes are packed into chromosomes that lie within each cell's nucleus. A chromosome is a protein-based structure that carries genomic information from one cell to the next.
Although genomics can apply to any organism, most of the attention has been on the human genome, which contains about 3 billion DNA base pairs, between 20,000 to 25,000 genes and 23 pairs of chromosomes. Genomics attempts to understand all the DNA and genetic material and advance the practical application of that knowledge.
Types of genomics
The scientific community is studying and experimenting with genomics for many different purposes. These various efforts have led to the emergence of multiple fields within the area of genomics, including the following:
- Structural genomics. Studies the entire genome to determine the structure of every protein encoded by the genome.
- Functional genomics. Collects and uses data from different genome projects, including sequencing projects, to describe gene and protein functions and interactions.
- Comparative genomics. Compares genome sequences in different species to gain a better understanding of their similarities and differences.
- Epigenomics. Studies the epigenetic changes in a cell's genetic material to understand how genetic changes can occur without altering the DNA sequence.
- Metagenomics. Analyzes the function and structure of complete nucleotide sequences from multiple organisms in a bulk sample, typically microbes, to better understand their inherent diversity.
- Pharmacogenomics. Studies how an individual's DNA affects the way that person responds to specific drugs, with the goal of providing patients with more effective treatments.
The term genomics is sometimes used interchangeably with genetics, but they are not the same. Genetics is concerned with how genes and their traits are inherited, while genomics looks at an organism's entire genome, including the genes and their interrelationships. Because genomics is concerned with the big picture, it can help identify the combined influence of all the genes on an organism's growth and development.
Genomics relies on DNA sequencing
To study an organism's genome, researchers must first conduct a process called DNA sequencing, which determines the exact order of nucleotide bases on a DNA strand. Only one strand needs to be sequenced because the nucleotide pairing determines the sequence on the second strand. Sequencing might also include the DNA found in the cell's mitochondria, rather than it's nucleus, or in its chloroplast if it's a plant.
Originally, DNA sequencing used analytical chemistry and molecule separation techniques to determine the order of the sequence. Scientists and technicians analyzed the sequences, which took a significant amount of time. However, sequencing techniques and equipment have advanced, making it possible to read DNA strands faster and in parallel. At the same time, sequencing instruments have become smaller, less expensive and more efficient.
The following techniques are just some of the methods scientists now use to sequence DNA:
- ChIP sequencing. Also called ChIP-seq, this approach identifies DNA-binding sites for transcription factors and other proteins through a combination of chromatin immunoprecipitation (ChIP) assays and massively parallel sequencing (MPS).
- Methylation sequencing. This sequencing method typically relies on bisulfite conversion to detect unmethylated cytosines in the DNA and convert them to uracils, making it possible to determine the percentage of methylated cytosines.
- Nanopore sequencing. In this sequencing method, single DNA strands pass through an electro-resistant membrane that contains tiny pores. Each pore includes a sensor that measures the electric current as the strand passes through. This information is then used to identify each nucleotide base and their exact sequence.
- Next-generation sequencing. This approach uses MPS technology to achieve high throughputs, making it possible to sequence an entire genome or targeted regions of DNA or RNA in a relatively short period of time.
- Sanger sequencing. Developed by two-time Nobel Prize-winning biochemist Frederick Sanger and his colleagues in the 1970s, their approach was the first method used for DNA sequencing. It was also the method used by the Human Genome Project. Sanger sequencing uses a chain-termination polymerase chain reaction to identify nucleotide bases.
- Targeted sequencing. This type of sequencing offers a quick and cost-effective approach for sequencing specific genomic regions with a high degree of accuracy, which is especially important in medical research. Several methods are available for targeted sequencing.
- Whole genome sequencing. This approach is used to sequence entire genomes. The process typically involves breaking down DNA into smaller segments that can be read by the sequencing machine. Each segment is also tagged so its position within the genome is identifiable.
Disk space estimates for storing human genomic data vary widely, ranging from 3 GB to 1 TB. The amount depends on how much raw and intermediate data might be needed to verify, refine and further analyze the data. The sequencing method and file types used also play a role in storage requirements.
Genomics in medicine and healthcare
In medicine, genomics and DNA sequencing can help medical professionals learn more about a patient's molecular biology. Genomic studies uncover the genetic makeup of patients, including their genetic differences and mutations. This information can be used to form a care plan specific to each patient's individual genetic composition, rather than treating the patient with a one-size-fits-all approach. Although still an emerging field, genomic medicine has the potential to inform all stages of healthcare, including prevention, diagnosis and treatment. It has gained the most traction so far in cancer, prenatal care, pharmacology, rare diseases and infectious diseases.
Next-generation genomic technologies make it possible to collect large amounts of genomic data. When this data is combined with bioinformatics, researchers can better understand drug responses and genetic-based diseases. This information can be used to achieve personalized or precision medicine, a model of healthcare in which providers customize treatment to fit each patient's needs and genetic configuration.
Early attempts at genome sequencing were slow and costly. The Human Genome Project was the first of its kind to completely sequence the human genome. The effort took 10 years and cost millions of dollars. A human genome can now be sequenced in less than 24 hours for under $1,000.
As genomic technologies have advanced, it has become easier and faster to collect genomic data. At the same time, researchers can better comprehend the data and its implications. The more refined these processes become, the more effectively healthcare providers will be able to use genomic information to diagnose and treat patients and create clinical decision support. Some pilot projects are attempting to integrate genomics capabilities into electronic health record systems.
Genomics is also playing a role in understanding genetic risk factors. Family health history can often reveal important risk factors for common and chronic diseases. Creating a genetic history is an important part of preventive medicine as well as public health. Perhaps one of the most widely used examples of this is noninvasive prenatal testing, also known by its acronym, NIPT. Since its introduction in 2011, NIPT uses next-generation sequencing to identify fetal chromosomal abnormalities early in pregnancy with a simple maternal blood test.
Experts believe that family history assessments offer several advantages. They can help lower the cost of providing care and lead to a better understanding of shared genetic and environmental risk factors. Performing genomic studies on family members can also provide further clues on their susceptibility to certain diseases, such as cancer or Alzheimer's disease.
Brief history of genomics
DNA was initially isolated as early as 1869, but it wasn't until the 1950s that the world saw the first technological advances, such as creating isotopes and radiolabel biological molecules. In 1953, scientists James Watson and Francis Crick described the structure of the DNA helix.
Modern genomics started in the early 1970s, when biochemist Frederick Sanger sequenced the first genomes using DNA from a virus and mitochondrion. Sanger and his team created techniques for sequencing, data storage, genome mapping and more.
Another scientist who played an important role in modern genomics is Walter Fiers. In 1972, he and his research team from the Laboratory of Molecular Biology of the University of Ghent in Belgium were the first to sequence a gene.
One of the most important developments occurred in 1990, when the National Institutes of Health and the U.S. Department of Energy launched the Human Genome Project. The project was a publicly funded international genomics research effort whose goal was to determine the sequence of the human genome and identify its genes. Project participants set out to sequence and identify all 3 billion DNA base pairs in the human genome.
The purpose of this project was to find the genetic roots of disease and help develop treatments. As part of their efforts, participants planned to make all human genome sequence information freely and publicly available within 24 hours of its assembly. The project successfully completed in April 2003.
Twelve years later, in early 2015, U.S. President Barack Obama announced a $215 million Precision Medicine Initiative, which aimed to tailor medical care to individuals based on their genes, lifestyles and environments. The National Cancer Institute received $70 million as part of the initiative to study cancer genomics.
"Over the years, innovations in sequencing protocols, molecular biology and automation increased the technological capabilities of sequencing while decreasing the cost, allowing the reading of DNA hundreds of basepairs in length, massively parallelized to produce gigabases of data in one run," according to a 2016 article in the journal Genomics.
Genomes evolve over time, changing in sequence or size. The study of genome evolution involves multiple fields and is constantly changing as more and more genomes are sequenced and made available to the scientific community and the public at large.
Genomic data testing has become more prevalent in medicine, which is making it even more important to enable it for electronic health record (EHR) integration. Explore why genomic data testing and EHR integration are so crucial.





